
Month: January 2025
Give Your Year to Our Lady – Crisis Magazine

The Church specifically places the Solemnity of Mary, Mother of God on the first day of the calendar year for a reason: our entire year should be given to the Blessed Mother.
PUBLISHED ON
January 1, 2025
The Solemnity of Mary, Mother of God is January 1st each year. The Church specifically places this commemoration on the first day of the calendar year for a reason: our entire year should be given to the Blessed Mother.
For too many Catholics, Mary is an afterthought. Devotion to her is too often seen as optional and only for the few priests and religious who desire to pray the Rosary. Catholics can view the role of Mary as superficial, unnecessary, or just plain plastic—not real life. She is fine to have in our artwork and portrayed in statues, but that is as far as it goes.
What we get wrong, even those who pray the Rosary regularly or who depend on Mary’s intercession, is that we are meant to have a relationship with Mary as Mother. Our conversations with her shouldn’t be built on formality or hiding behind built up piety. Like any great mom, she desires for us to have real and raw conversations with her.
Orthodox. Faithful. Free.
Sign up to get Crisis articles delivered to your inbox daily
Sign up to get Crisis articles delivered to your inbox daily
My own journey with Mary has intensified this year because my own mother has been struggling in health. In her constant doctors’ visits and especially in her hospitalization, I couldn’t stop talking to and thinking of the Blessed Mother. Ever since my mom has been battling serious illness, I have had a renewed understanding of just how much I love her and need her.
There is nothing like the relationship between a mom and her child. The ability to talk to her about anything and the peace that comes from simply being with her is unparalleled. When we reflect on the relationship of Mary and Jesus, we will find that their intimacy is what we are invited into. The beautiful intensity that I feel in my relationship with my mother during this time is a glimmer of what Mary and Jesus had. It is not an optional part of the spiritual life but a needed aspect for seeing and knowing Christ.
This spiritual application of the mother-child relationship to Mary can be seen in Pope St. John Paul II’s papal motto of “totus tuus.” This meant that John Paul was saying his whole life was “totally yours”—totally Mary’s.
The future pope’s mother died when he was a teenager. Following her death, it is reported that his father took him to a local church, and as they prayed before a shrine of Mary, he told his son, “This is your Mother now.” The pope took this call seriously. He saw that the closer he got to Mary, the more he was able to see, know, and trust her Son. She gives us access to see Jesus through her own eyes.
Our Lady knew what suffering was. So, when we suffer and those around us suffer (like my mom), we have the deep surety that all circumstances will be resurrected. When you suffer in 2025, turn to Mary and see the cross through her eyes. Learn to love Christ more by being more devoted to His Mother.
Our Lady knew what suffering was. So, when we suffer and those around us suffer, we have the deep surety that all circumstances will be resurrected. Tweet This
Our Lady knew what it was like for the future to be known. She also knew, in the midst of everything, that God was powerfully with her. This is the beauty of dedicating our whole year to Mary: she never fails in bringing us to her Son. She exists to reflect the glory of God made man.
The future that is before us is unknown. There might be uncertainties about your own health or financial situation. There might be fears and anxieties about our country or Church. No matter what our concern is over the unknown, we can depend on Mary to guide us through. For she was betrothed and was found pregnant. She had no place for her child to be born. She had to flee to a foreign country because Herod was trying to kill her Son. She was told in the Temple that a sword would pierce her heart.
Through it all, she kept Christ at the center. Through it all, she can aid us in doing the same. For the entire reason for the Christian life is to know, love, and serve the Lord. So, this year, go to the one who knows Him best so that you can meet Him in this life and in the one to come.
As Pope St. John Paul II said in a homily on today’s solemnity in 2002, “If Jesus is Life, Mary is the Mother of Life. If Jesus is Hope, Mary is the Mother of Hope. If Jesus is Peace, Mary is the Mother of Peace, Mother of the Prince of Peace.” Our Lady gave birth to true life, hope, and peace. Our Lady gave birth to God. She bore Christ in her womb, and she will do all in her power so that faith can be born in us.
All we have to do is give her permission to do so.
She’s waiting for you. Give her permission to change your year and your life.
Thomas Griffin is the chair of the religion department at a Catholic high school on Long Island where he lives with his wife and two sons. He is the founder and editor-in-chief of Empty Tomb Project: The Magazine.
Comments are a benefit for financial supporters of Crisis. If you are a monthly or annual supporter, please login to comment. A Crisis account has been created for you using the email address you used to donate.
There are no comments yet.
Published on
Copyright © 2025 Crisis Magazine. All rights reserved.
Created by Perceptions Design Studio.
Lost your password?
Use a social account for faster login or easy registration.
PredictWind 2024 Moth Worlds: Matthias Coutts leads Oceania Championship after Day 1 – Sail World

Local sailor Matthias Coutts leads the 2024 PredictWind Moth Oceania Championship after the first day of racing.
Sailed at Manly Sailing Club, about an hours drive north of Auckland, 58 competitors contested racing in the Pre-Worlds event, which was sailed with rain squalls passing regularly through the race fleets.
Four races were sailed across the Blue and Yellow fleets, with only one competitor, Leonard Takahasi (NZL) finishing the fourth race in the Blue fleet.
Classified as a Youth sailor, but racing in Open competition Coutts battled with fellow Manly and Youth sailor, Jacob Pye in the four races sailed in the Blue fleet. At the end of the first days racing the are just 1pt apart on the overall results. Pye was unlucky to miss being awarded the 2023 World Championship title after being a clear leader in the Worlds after two races. Due to light winds at Weymouth UK, no further racing was possible and the world championship was declared a no-contest.
Between them Coutts and Pye won three of the four races sailed in the Blue fleet
USA sailor Riley Gibbs, from the American Magic America’s Cup team is third overall 9pts behind Matthias Coutts.
US sailors round out the top five overall, with Richard Didham on 21pts and a second American Magic team member Harry Melges, also a Youth sailor on 25pts.
Sailing’s rockstars are scattered well down the fleet, with Paris 2024 Olymnpic Gold medalist and 2024 SailGP champion Diego Botin (ESP) in 24th overall. American Magic helmsman Lucas Calabrese (USA) is 37th overall and French America’s Cup helmsman Kevin Peponnet is 48th overall after only competing in two races.
Racing continues on Thursday.
Full results can be found here.
Move Over SOL and ETH: These 5 Altcoins Are the New Crypto Powerhouses – Analytics Insight
Every Movie Starring Kevin Costner On Netflix – Screen Rant
Hong Kong group gives government failing grade on efforts to help children – South China Morning Post

Children’s Rights Association gives score of 26 out of 100, naming housing as their biggest concern, followed by medical services and education for 2025
The Children’s Rights Association on Wednesday released the report for 2024 evaluating authorities’ performance in legislation, policies and services targeting poor children and their families.
While 26 is below the pass mark of 50, the latest score was the highest since the group started releasing its annual review in 2005. The association gave a score of 21 in 2023 and 19 in 2022.
The rating was decided by more than 20 children ambassadors, aged 17 and under, and after consulting with about 5,000 other members, all minors, through its app and various meetings.
“The government has not made much progress on its policies addressing social challenges such as poverty, the wealth gap, medical care and welfare,” said Lau Yui, one of the ambassadors.
The 11-year-old Primary Six pupil said the average waiting time for public housing remained long and little policy progress had been made in areas such as healthcare, poverty alleviation or legislation banning discrimination against new arrivals from mainland China.
“The current government focused on economic recovery but neglected to comprehensively tackle the problem of child poverty through policies, legislation or services,” he added. “Its work progressed slowly and could not keep up with social needs.”
From 'worst president' to 'highest respect': Trump softens opinion of Jimmy Carter in death – USA TODAY

WASHINGTON – President-elect Donald Trump had only kind words to say about Jimmy Carter upon his death on Sunday, calling the former president “a truly good man” who will be missed.
At Mar-a-Lago, Trump’s estate in Palm Beach, Florida, flags were flying at half-staff on Monday in Carter’s honor.
Trump offered a far less charitable view of Carter when he was alive.
For years, Trump, a Republican, has mocked the one-term, Democratic commander-in-chief as the nation’s worst president. Just two months ago, on Carter’s 100th birthday, Trump suggested the terminally ill centenarian was happy because Joe Biden had finally replaced him as the worst president ever.
Carter often gave as good as he got. A Georgia peanut farmer before he got his start in politics, Carter was no Georgia peach when it came to his assessment of Trump.
When late-night TV host Stephen Colbert asked him in 2018 if Americans wanted “kind of a jerk” as their leader, Carter didn’t miss a beat. “Apparently, from this recent election, yes,” he retorted.
Earlier this year, Carter suggested to his son, Chip Carter, that he wanted to live long enough to vote for Trump’s rival, Vice President Kamala Harris, in November. Two weeks before election, Carter’s office confirmed that he had voted by mail. For Harris.
Amid the barrage of insults and smack talk were temporary moments of rapprochement between the 39th and the 45th presidents.
During Trump’s first term, Carter occasionally came to his defense.
Carter not only attended Trump’s first inauguration, he was the first former president to RSVP. Later that year, in 2017, Carter suggested the media had been harder on Trump than any other president. Trump responded by tweeting a message of thanks to Carter for “the nice remarks.”
Jimmy Carter on long life, philanthropy:‘I’ve had an exciting and adventurous and gratifying existence’
In 2019, Trump’s White House announced that Carter had written Trump a “beautiful” letter about trade talks with China and that the two men had had a “very good telephone conversation” on China and other topics.
“The president has always liked President Carter and First Lady Rosalynn Carter and extended his best wishes to them on behalf of the American people,” the White House statement said.
If Trump liked Carter, he seldom let it show.
In 2014, Trump joked that Carter was dead, even though the former president was very much alive. While speaking at the Conservative Political Action Conference, Trump referred to the “late, great Jimmy Carter.”
“Of course I don’t think Jimmy Carter is dead,” he tweeted later amid criticism for the gaffe. “Just being sarcastic,” he said, adding, “but never thought he was alive as President, stiff!”
On several occasions, Trump called Carter the worst president in history – at least until Barack Obama came along.
“I never thought I’d say it in my lifetime, but President Barack Hussein Obama, aka Barry Sotoro, is a far worse president than Jimmy Carter!” Trump said in 2014.
Trump, who has a tendency to change his stories to suit his audience, offered a slightly different take after Biden defeated him in 2020. He suggested several times that Biden, not Obama, had replaced Carter as the worst president ever.
Jimmy Carter state funeral:President to be honored in Washington before burial in Georgia
For his part, Carter called Trump “a disaster” of a president during a 2018 interview with The Washington Post and suggested the Republican held “an attitude of ignorance toward the truth.”
The next year, Carter told his church congregation in Plains, Georgia, that Trump had called him to discuss the letter he had sent to Trump about China. Carter suggested China would likely surpass the U.S. as an economic superpower because it had been able to avoid the military entanglements that have drained funds that might otherwise have been spent on infrastructure.
“I don’t really fear that time, but it bothers President Trump, and I don’t know why,” Carter said. “I’m not criticizing him – this morning,” he added.After Trump was elected to a second term in November, Carter, who had been in hospice for nearly two years, announced he would not attend Trump’s inauguration but would be there if not for his deteriorating health.
In a pair of statements after Carter’s passing, Trump made reference to his disagreements with the former president.
This time, however, he not only dispensed with the insults, he even suggested that Carter had earned his respect.
“While I strongly disagreed with him philosophically and politically, I also realized that he truly loved and respected our Country, and all it stands for,” he wrote. “He worked hard to make America a better place, and for that I give him my highest respect.”
Michael Collins covers the White House. Follow him on X @mcollinsNEWS.
Multiple security vulnerabilities detected in Chrome desktop browser: CERT-In – Deccan Herald
Three Members Form a Nascent Religious Community – The Living Church

Haven Religious is an emerging religious community in Hartford, Connecticut that hopes to provide active service to the church and the city. There are three members: the Rev. Marta Rivera Monclova, Gregory Simmons, and Sean Glenn. Rivera Monclova and Simmons are novices in the Order of the Body of Christ, a traditional vowed religious community that exists within Haven.
Rivera Monclova is the leader. She wants to create something she sees as not prominent within the Episcopal Church: active religious life. “We’re very inspired by the early Victorian sisterhoods, who often were just women who started living together in a house and serving the church,” she said.
The community uses the Carmelite Rule of St. Albert, because it was written for a group of hermits who wanted to unite into a more corporate life. Because members can pursue different ministries, as opposed to having one shared apostolate, they felt like a rule emphasizing individual discernment and spirituality would work best. The community shares in daily Morning Prayer and Evening Prayer, acknowledging that sometimes ministry needs mean someone cannot attend.
Haven has been welcomed by the Episcopal Church in Connecticut. Haven chose Connecticut partly because it has a canon dedicated to intentional communities. The Episcopal Church requires six professed members before it recognizes a community as a religious order. Members found it appealing that Connecticut has regulations for smaller communities.
Rivera Monclova, who was ordained to the priesthood in January, is a curate at Grace Episcopal Church in Hartford. Simmons is assistant to the dean and event coordinator at the cathedral, and Glenn is its office assistant. One of Rivera Monclova’s dreams is for the community to provide supply service not just for preaching and the Eucharist, but for a parish’s other needs.
Grace Church recently distributed free turkeys through its food pantry, and if two additional people had helped, it would have made a big difference. This could be especially useful to parishes that struggle financially. “I’m seeing it more and more: the areas of the church that most need help and service are the least able to pay for it,” she said.
This financial struggle also affects younger clergy, especially in Connecticut, where many positions are only half- or quarter-time. Haven wants to provide an inexpensive residence for young priests. Priests might then decide whether to join the Order of the Body of Christ. Glenn is discerning whether he wants to become a vowed member, but whatever he decides, he can keep living there and remain part of the community.
“There’s a hunger among young people of faith for a residential Christian community,” Rivera Monclova said. She says Episcopal Service Corps it does a good job of catching young people on fire for ministry but then does not give them anywhere to take that fire besides ordination. “Surely we’re not saying the only call of dedicated service to the church is the priesthood,” she said.
While Haven more broadly welcomes everyone, the Order of the Body of Christ, is intended to be a vowed order following the evangelical counsels of poverty, chastity, and obedience. “We want to have traditional vows for non-traditional people,” Rivera Monclova said.
She considered existing women’s communities within the Episcopal Church, but decided they weren’t for her. “I visited one when I was 42, and I would’ve been the youngest sister by 20 years. That’s a calling somebody could have, but it’s not one I had. I’d still be starting something new, but in a container of something older, and I felt like that container would cause more friction than energy,” she said.
She wasn’t interested in joining a dispersed order without a discipline of celibacy because she wanted to be fully committed to service to the church and the community. “It’s a lens through which you see the world, when you have one person that’s your person. It’s why we take marriage so seriously. Even having it as an option would be a distraction,” she said.
“I want to be in a community where we’re all facing the same direction,” she added, comparing it to ad orientem worship, in which both congregants and the celebrant face the altar.
Haven is an Anglo-Catholic community. Simmons is helping coordinate Eucharistic adoration at Grace Episcopal Church during Advent. Haven members pray the Angelus corporately, but they modernize the language. But Anglo-Catholicism’s heritage isn’t just about chanting and incense but also about service to the poor.
Rivera Monclova hopes to begin celebrating the Eucharist for Church by the Pond, the Cathedral’s outdoor ministry that primarily serves homeless people. She also helps run Place of Grace Food Pantry at her parish. “The need in Hartford is huge,” she said. Before the COVID-19 pandemic, Grace’s food pantry served 100 to 150 households. Since then, it has risen to 350 households, and has given out 427 turkeys.
There are many direct service organizations in Hartford, and Haven wants to help them instead of creating new groups. On the other hand, secular social service organizations can’t provide for people’s spiritual lives. “Some of the people who are coming are hungry for more than bread,” she said. “Who around the parish has spiritual needs we could be addressing that we’re not?” She acknowledged that asking strangers what they want in a church is a particularly challenging form of congregational development.
Hartford is also home to Trinity College, which is historically affiliated with the Episcopal Church. Rivera Monclova has officiated at the college’s choral Evensong, and she hopes that as the community develops, there can be further partnerships between Haven and the college, especially with the schola.
Greta Gaffin is a freelance writer based in Boston. She has a master of theological studies degree from Boston University and a bachelor’s degree in economics from the University of Massachusetts Amherst.
Top headlines. Every Friday.
© 2024 The Living Church Foundation. All rights reserved.
KORE-Map 1.0: Korean medicine Omics Resource Extension Map on transcriptome data of tonifying herbal medicine – Nature.com
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
Advertisement
Scientific Data volume 11, Article number: 974 (2024)
787
Metrics details
Traditional herbal medicine, rooted in a long history of use in East Asia, combines several herbs to create treatments showing high efficacy with minimal side effects, for specific diseases. Such combination therapies represent a potential reservoir of new drugs for treating multifactorial and incurable chronic diseases. However, the complexity of their mechanisms of action due to the combination of multiple compounds, has limited their research integration into modern pharmacological science. To address this challenge, we constructed drug-induced transcriptome data for herbal medicines through systematic experiments, analyzed with the aid of various omics databases. We introduce KORE-Map 1.0 (Korean medicine Omics Resource Extension Map), the first comprehensive resource of drug-derived transcriptome data for representative tonifying herbal medicines, effective in enhancing the immune system. This dataset aims to provide novel insights into the combinatorial mechanisms of these herbal medicines and to aid in the discovery of new therapeutic targets and indications for various incurable diseases.
Herbal medicine has long been used effectively for disease treatment in East Asia, notably in Korea and China1. These medicines exemplify combination therapy, employing multiple compounds to treat diseases2. Such therapy is effective in multifactorial diseases as it addresses multiple targets3, and offers the advantage of fewer side effects such as drug resistance4. Tonifying herbal medicine (THM), a form of combination therapy, not only targets diseases directly but also aims to activate immunity5. Thus, THMs have the capacity to: (1) treat multifactorial diseases, (2) minimize drug resistance, and (3) manage conditions challenging to treat with conventional drugs by bolstering the immune system. This study therefore proposes a new paradigm in disease treatment.
Despite its numerous benefits, research on THM faces limitations, particularly in accurately identifying the mechanism of action (MOA) due to the involvement of multiple compounds. One solution is drug-induced transcriptome analysis6,7, a method of measuring relative expression levels of mRNA after treating a cell line with a specific drug. This method reveals the MOA of drugs through complex pharmacodynamic processes as reflected in mRNA expression8, providing insights into the therapeutic effects of both single drugs and compound combinations. Furthermore, it facilitates the identification of transcriptomic signatures responsive to these treatments, confirming the therapeutic mechanism of each approach. Therefore, transcriptome analysis, by elucidating the MOA of combination therapies, is pivotal in revealing the therapeutic mechanisms of herbal medicine prescriptions beyond individual herbs.
To uncover the therapeutic MOA of herbal medicines using transcriptome analysis, it is important to consider a variety of factors. One such variable to consider is the choice of cell line9. As the dominantly expressed genes vary across cell lines, analysis across diverse cell lines essential for confirming the MOA of herbal medicines on multiple targets. Therefore, generating transcriptome data from various cell lines is an efficient strategy that not only validates the therapeutic effects of herbal medicines, but also elucidates their mechanisms of action in various organs. Another critical factor is drug concentration, which can significantly influence the MOA of drugs10. For example, Finasteride, a well-known treatment for benign prostatic hyperplasia, demonstrates different effects at varying doses—acting on prostate cancer at higher doses (5 mg/day), and delaying hair loss at lower doses (1 mg/day)11. This variability demonstrates the importance of dose determination in confirming the MOA of herbal medicines. Additionally, the choice of solvent plays a pivotal role7, as natural products comprise both hydrophilic and hydrophobic compounds. Therefore, the fractions of compounds extracted from water and ethanol solvents vary, indicating different processing mechanisms. To reveal the therapeutic mechanisms of herbal medicine, multiple variables must be considered. Transcriptome data, produced in consideration of these variables, are important in elucidating the MOA of the drug.
In this Data Descriptor, we introduce KORE-Map 1.0 (Korean Medicine Omics Resource Extension-Map), featuring THM-derived transcriptome data available on the NCBI gene expression omnibus (GEO) platform. The data were generated from four THMs commonly used in clinical practice, along with the 10 herbs constituting them. The transcript expression information was derived from both water and ethanol extracts of THMs and herbs, prepared at three different concentrations and applied to four representative human-derived cell lines (A549, HepG2, HT29, and SW1783). Utilizing the MGIEasy RNA directional library prep kit and the MGISEQ-2000 sequencing system, both widely used worldwide, not only facilitates easy data reuse but also ensures excellent compatibility with other transcriptome datasets. The THM-derived transcriptome data produced in this study could serve multiple purposes, such as aiding in the identification of therapeutic MOAs involving multiple compounds, which is crucial for understanding therapeutic mechanisms in multifactorial or comorbid conditions. In addition, the transcriptome data, spanning various cell lines and concentrations, hold potential for applications in drug repositioning, side effect detection, and more, by enabling the simultaneous evaluation of the effects of multiple compounds on multiple targets and organs.
Dried medicinal plants, conforming to the Korean Pharmacopoeia standards, were provided by Kwangmyung-dang Medicinal Herbs Co., located in Ulsan, Republic of Korea. These samples underwent an organoleptic examination by Dr. Choi Goya, a herbal medicine organoleptic examination expert appointed by the Korea Food and Drug Administration. The identification to species level was accomplished through DNA barcode region sequencing. Voucher specimens have been stored at the Korean Herbarium of Standard Herbal Resources, within the Herbal Medicine Resources Research Center, at the Korea Institute of Oriental Medicine in Naju, Republic of Korea (Table 1). All herbs and extracts, which were sourced from the Oriental Medicine Resources Research Center (KIOM), are available online at https://oasis.kiom.re.kr/herblib.
Hot water and 70% ethanol extracts of each plant were prepared and supplied by KOC Biotech Co., located in Daejeon, Republic of Korea. Initially, dried plants (1,000 g) were pulverized and extracted in 10 L of hot distilled water for 3 h using a reflux extraction system (MS-DM609; MTOPS, Seoul, Republic of Korea), or in 10 L of 70% ethanol for 1 h using an ultrasonication system (VCP-20, Lab companion, Dajeon, Republic of Korea) twice. The resulting extract solutions were filtered through a 5 µm cartridge filter, concentrated using a rotary evaporator (Ev-1020, SciLab, Seoul, Republic of Korea), and finally lyophilized in a freeze dryer (LP-20, Ilshin-Bio-Base, Dongducheon, Republic of Korea) to produce the final extracts. These extracts were then finely homogenized and packaged in glass bottles with desiccant silica gel. THMs were prepared by blending and homogenizing these extracts in accordance with the composition ratios and extract yields of the individual medicinal herbs, according to the Korean Pharmacopoeia (Table 2). For in vitro applications, extracts (100 mg) were vigorously vortexed for 30 min in 10 mL of phosphate-buffered saline (PBS; Thermo Fisher Scientific, Rockford, IL, USA) containing 2% DMSO. This mixture was then sterilized by filtration through a 0.22 µm membrane to obtain a stock solution (10 mg/mL), which was divided into small aliquots and stored at −80 °C until their use.
All cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA) and were cultured in a basal medium enriched with 10% heat-inactivated fetal bovine serum, 100 IU/mL penicillin, and 100 µg/mL streptomycin, all within a humidified incubator (Table 3). Cell confluence levels between 80–90% prompted the replacement of the growth medium every 3–4 days to maintain optimal growth conditions. To ensure the absence of mycoplasma contamination, the MycoAlert PLUS mycoplasma detection kit (Lonza, Rockland, ME, USA) was employed for regular testing.
To determine the appropriate treatment drug concentrations, we performed cell cytotoxicity tests to investigate drug doses that maintained 80% cell viability (IC20s), which were then adopted as the maximal doses for RNA-seq data collection. For drugs whose IC20s could not be determined, the highest treatment concentrations were capped at 500 µg/mL for extracts, considering both their solubility and relevance for clinical application. To confirm the influence of concentration, cells were treated with three different concentrations using 1/5 serial dilutions, thereby exposing them to low, medium, and high doses. Positive control drugs such as wortmannin (Sigma, W1628), LY294002 (Sigma, L9908), and Thioridazine (Sigma, T9025) were incorporated into the assay for comparative analysis against the connectivity map (CMap) data. Cells treated with a 2% DMSO/PBS solution served as the vehicle control. One day before drug administration, cells were seeded into 6-well culture plates with 3 mL of growth medium. Following a 24 h treatment period, the cells were washed with ice-cold PBS, and total RNA was isolated using QIAzol RNA isolation reagents (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Total RNA (over 500 ng) from each sample was processed for the mRNA sequencing library using the MGIEasy RNA directional library prep kit (MGI Tech Co., Ltd., China), following the manufacturer’s instructions. The library concentration was quantified using the QuantiFluor® ssDNA System (Promega Corporation, WI, USA). The prepared DNA nanoball was sequenced on an MGISEQ system (MGI Tech Co., Ltd., China) employing 100 bp paired-end reads. The RNA-seq data quality was assessed using FastQC (v0.11.9). To remove common MGISEQ adapter sequences, TrimGalore (v0.6.6) was utilized. Trimmed reads were then mapped to the human reference genome assembly GRCh38 (hg38) using the STAR aligner (v2.7.3a) with default settings12. Gene transcript abundance, including expected read counts and transcripts per million, was quantified using RSEM (v1.3.3), with the gene annotation GRCh38.8413. The raw sequence data (FASTQ files) and the preprocessed expression values for each gene have been deposited in the GEO under accession numbers GSE244687, GSE244707, GSE244694, and GSE245912.
Using the gene symbols of protein-coding genes, we utilized the collapseRows function from the WGCNA package (v.1.72-1)14, specifically designed to merge expression data for genes represented by multiple probes. This approach effectively reduces redundancy and potential noise, enhancing the clarity of subsequent analyses. Additionally, the filterByExpr function from the genefilter package (v.1.78.0)15 was utilized to exclude genes that failed to meet predetermined expression criteria across samples. This filtering ensured that only genes most likely to provide reliable and relevant signals were retained for analysis.
For evaluation of each set of treatment conditions—encompassing four cell lines, 14 herbs and THMs, two extraction methods, and three concentration levels— we conducted differential gene expression (DGE) analysis against the corresponding control samples. This analysis was performed using the Wald test statistic as implemented in the DESeq. 2 package (v.1.36.0)16. Differentially expressed genes (DEGs) were determined based on a fold-change threshold of 1.5 and an adjusted P-value of less than 0.05.
The fold-change values derived from the DGE analysis across all treatment conditions were clustered using the t-distributed stochastic neighbor embedding (t-SNE) algorithm. This machine learning technique, designed for dimensionality reduction, excels in visualizing high-dimensional datasets, making it a valuable tool for interpreting complex gene expression patterns. The analysis was conducted utilizing the Rtsne package (v.0.16), with the perplexity parameter set to 1017.
Connectivity Map data were obtained from the Clue.io platform(clue.io/data/CMap2020#LINCS2020). For our analysis, we selected level five gene expression signatures with high reproducibility, defined by moderated z-scores that met specific criteria (distil_cc_q75 > 0.5 and pct_self_rank_q25 > 0.05), to compare with our RNA-seq data. The R package CMapR (v1.8.0) was used to manipulate the level 5 GCTX file (level5_beta_trt_cp_ n720216 × 12328.gctx). Given the variance in gene expression profiling between our RNA-seq data and L1000 assays9 used in CMap, a direct comparison between gene expression values was difficult due to distinct distributions of expression values. To navigate this, we employed gene set enrichment analysis (GSEA) as an alternative method to explore the genome-wide perturbing effects of treatments such as wortmannin at the pathway level18. We utilized 2,229 gene sets from several databases—Hallmark, Biocarta, KEGG, REACTOME, PID, and Wikipathways—available through MSigDB (https://www.gsea-msigdb.org/gsea/msigdb). The analysis involved performing GSEA on all genes, ranked according to their Wald test statistics or level5 z-scores. To obtain the MSigDB gene sets and conduct GSEA, we utilized the R package MSigDBR (v7.5.1) and FGSEA (v3.18). From the GSEA results, we defined pathway activity score (PAC) as ‘sign (enrichment score) × -log10(p-value)’ value to quantify the significance level. PAC vectors of equal lengths (n = 2,229) were generated for both our dataset and the CMap dataset. Subsequently, we determined the Pearson correlation coefficient to assess the relationship between the PACs from our samples and those from CMap (Fig. 1).
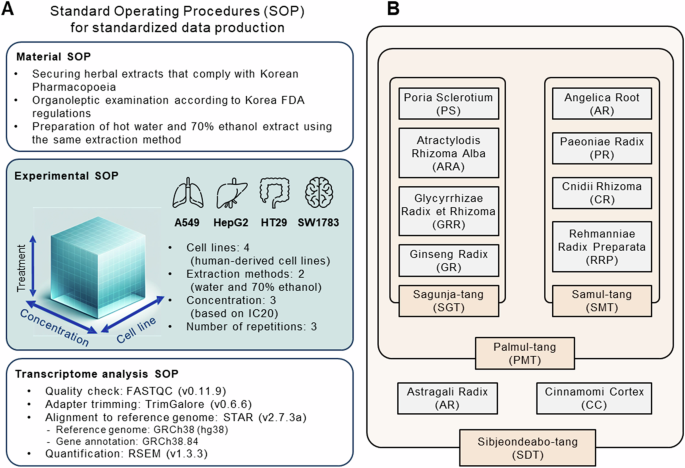
Introduction to transcriptomic data production protocols and herbal drugs used. (A) Overview of standard operating procedures for producing standardized transcriptome data. (B) List of herbal drugs processed for transcriptome data production.
All raw transcriptome data were uploaded to GEO in the FASTQ format using paired-end sequencing files. Each data file was presented in two fq.gz formats. Essential details such as the production method, adopted cell line, and dosage information were included in the metadata accompanying the GEO submission19,20,21,22 (Table 4). The dataset submitted to GEO comprised 1,092 RNA sequencing samples across 21 batches (Table 5, Supplementary Tables 1–6). Transcript samples were derived from four distinct cell lines: 270 (A549; accession number, GSE24468719), 270 (HepG2; accession number, GSE24468720), 273 (HT29; accession number, GSE24468721), and 279 (SW1783; accession number, GSE24468722). The difference in data volume between HT29 andSW1783 cell lines can be attributed to two factors:(1) A discrepancy in drugs used as positive controls, and (2) variations in the number of transcript production batches due to technical issues. Wortmannin, known for its anti-inflammatory properties, served as a universal positive control across all cell lines. Further, LY294002 produced in HT29 cells, and LY294002 and Thioridazine produced in SW1783 cells served as additional positive controls, contributing three and six samples, respectively. Consequently, six batches were specifically allocated for the SW1783 cell line, with an inclusion of three extra samples.
To ensure the suitability of samples for downstream sequencing, RNA quality and integrity were thoroughly evaluated. The optical density at 260 and 280 nm was measured using the Trinean Dropsense™96 micro-volume reader. The A260/A280 ratio serves as an estimate of RNA purity, with values exceeding 1.8 indicating relatively high purity. Our analysis revealed that the RNA samples typically exhibited a ratio close to 1.8, signifying a substantial proportion of pure RNA (Fig. 2a). Furthermore, the 28S/18S rRNA ratio and the RNA integrity number (RIN) were measured using an agilent bioanalyzer DNA chip to assess the extent of RNA degradation. All RNA samples demonstrated a 28S/18S ratio approximately equal to 2 and an RIN value of 7 or above, reflecting high RNA quality and integrity (Fig. 2b). These results suggest that the RNA is of suitable quality for downstream RNA sequencing23.
Quality assessment of RNA samples. (A) The A260/A280 ratio for individual samples grouped by four cell lines. The minimum of values widely recognized as indicative of high purity RNA is represented by the dotted line. (B) The 28 s/18 s rRNA ratio (left) and RNA integrity number (right) for individual samples grouped by four cell lines. Each dotted line represents a minimum value widely known to reflect high RNA quality and integrity.
The quality of the raw RNA-seq data was assessed using FastQC (v0.11.9), a software that generates a detailed report, including metrics such as per-base quality scores and GC content distribution. A representative FastQC report indicated that the overall read quality was high (Fig. 3b). Similar quality metrics were observed in all other FastQC reports, qualifying them for further analysis. To obtain clean data, adapter sequences and low-quality bases (Phred score below 20) were removed using TrimGalore (v0.6.6). As a result, a high percentage of reads, with a median of 96.67%, were successfully and uniquely mapped to the human reference genome GRCh38 (hg38) (Fig. 3b)24.
Quality evaluation of RNA sequencing data. (A) Representative FastQC report showing per sequence quality scores (left) and GC content (right) for A549 cell line treated with dimethyl sulfoxide (DMSO). (B) Summary of unmapped, multiple-mapped, and uniquely mapped reads against the reference genome for each cell line.
To ensure the reproducibility of our RNA-seq data, we quantified biological and technical batch effects by analyzing expression levels (TPM values for 19,826 protein-coding genes). Initially, the biological reproducibility was assessed through the analysis of three independent biological replicates for each treatment condition; cell line, treatment, and dose. Each replicate involved separate RNA extraction, RNA-seq library preparation, and sequencing processes. We calculated the pairwise Pearson’s correlation coefficient among replicates to quantify their similarities. This revealed a high degree of biological reproducibility, with an average correlation coefficient of 0.994 across all conditions. Furthermore, 97.8% of the conditions exhibited an average expression level correlation exceeding 0.95 across the three replicates (Fig. 4a).
Replicability of RNA-seq profiles. (A) Distribution of Pearson’s correlation coefficients for replicates (yellow) versus different samples (gray). (B) Heatmap of Pearson’s correlation coefficients among replicate samples across various sequencing batches.
Technical reproducibility was subsequently evaluated to address potential batch effects arising from sequencing. Since a single sequencing lane can accommodate up to 60 samples, we distributed samples from the same cell line across six different sequencing batches (A to F). Control samples treated with the vehicle were included and sequenced in all six batches, to assess batch effects. The correlation coefficients between control samples from different batches were calculated, indicating minimal batch effects. Notably, all control samples exhibited high correlation coefficients (>0.99) with samples sequenced in different batches (Fig. 4b).
To verify the reliability and reproducibility of our RNA-seq data, we compared our drug-induced transcriptome profiles to those generated by the CMap 9, a comprehensive database featuring gene expression profiles of human cell lines treated with various bioactive compounds. We chose wortmannin, an established positive control that is also included in the CMap dataset, as a benchmark for our analyses across three cell lines: A549, HEPG2, and HT29.
To facilitate direct comparison between transcriptome data generated from different platforms, we converted gene-level expression values to pathway-level scores. This approach aggregates the expression changes across genes within 2,229 well-defined biological pathways, providing a more robust and interpretable measure of pathway activation or inhibition.
We then compared the pathway-level scores resulting from wortmannin treatment in our study with those generated by CMap. The pathway-level scores from our wortmannin treatment analysis showed a high correlation with those obtained from CMap (Fig. 5). This notable concordance serves as strong evidence of the reliability and reproducibility of our RNA-seq data, affirming its ability to capture drug-induced changes in cellular pathways.
Transcriptome data comparisons with CMap. Distribution of Pearson’s correlation coefficients for samples under the same treatment condition (yellow) versus different conditions (gray).
All software used to analyze the RNA-seq data, along with their parameters are clearly described in the Methods section. Unless specified otherwise, default parameter settings recommended by the developers were used. The curation and validation of the dataset were conducted using custom R code, as detailed in the Materials and Methods section. The source code specifically employed for preprocessing the RNA-seq data is publicly available through the GitHub repository (https://github.com/LeeLab-Sysbio/KOREMAP.v1). Researchers are encouraged to cite this paper when utilizing the RNA-seq data uploaded in GEO.
Tyler, V. E. Herbal medicine: from the past to the future. Public Health Nutr 3, 447–452, https://doi.org/10.1017/s1368980000000525 (2000).
Article CAS PubMed Google Scholar
Gouws, C., Steyn, D., Du Plessis, L., Steenekamp, J. & Hamman, J. H. Combination therapy of Western drugs and herbal medicines: recent advances in understanding interactions involving metabolism and efflux. Expert Opin Drug Metab Toxicol 8, 973–984, https://doi.org/10.1517/17425255.2012.691966 (2012).
Article CAS PubMed Google Scholar
Rosini, M. Polypharmacology: the rise of multitarget drugs over combination therapies. Future Med Chem 6, 485–487, https://doi.org/10.4155/fmc.14.25 (2014).
Article ADS CAS PubMed Google Scholar
Basanta, D., Gatenby, R. A. & Anderson, A. R. Exploiting evolution to treat drug resistance: combination therapy and the double bind. Mol Pharm 9, 914–921, https://doi.org/10.1021/mp200458e (2012).
Article CAS PubMed PubMed Central Google Scholar
Leung, H. Y. & Ko, K. M. Differential Effects of Yin-and Yang-Chinese Tonifying Herbs on Innate and Adaptive Immunity. Chinese Medicine 14, 68–78 (2023).
Article CAS Google Scholar
Baek, S. J. et al. Identification of a novel anticancer mechanism of Paeoniae Radix extracts based on systematic transcriptome analysis. Biomed Pharmacother 148, 112748, https://doi.org/10.1016/j.biopha.2022.112748 (2022).
Article CAS PubMed Google Scholar
Park, S. M. et al. Systematic transcriptome analysis reveals molecular mechanisms and indications of bupleuri radix. Front Pharmacol 13, 1010520, https://doi.org/10.3389/fphar.2022.1010520 (2022).
Article CAS PubMed PubMed Central Google Scholar
Lee, H., Kang, S. & Kim, W. Drug Repositioning for Cancer Therapy Based on Large-Scale Drug-Induced Transcriptional Signatures. PLoS One 11, e0150460, https://doi.org/10.1371/journal.pone.0150460 (2016).
Article CAS PubMed PubMed Central Google Scholar
Subramanian, A. et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 171, 1437–1452 e1417, https://doi.org/10.1016/j.cell.2017.10.049 (2017).
Article CAS PubMed PubMed Central Google Scholar
Waldmann, T. et al. Design principles of concentration-dependent transcriptome deviations in drug-exposed differentiating stem cells. Chem Res Toxicol 27, 408–420, https://doi.org/10.1021/tx400402j (2014).
Article CAS PubMed PubMed Central Google Scholar
Libecco, J. F. & Bergfeldt, W. F. Finasteride in the treatment of alopecia. Expert Opin Pharmaco 5, 933–940, https://doi.org/10.1517/14656566.5.4.933 (2004).
Article CAS Google Scholar
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, https://doi.org/10.1093/bioinformatics/bts635 (2013).
Article CAS PubMed Google Scholar
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinformatics 12 https://doi.org/10.1186/1471-2105-12-323 (2011).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. Bmc Bioinformatics 9 https://doi.org/10.1186/1471-2105-9-559 (2008).
Gentleman, R., Carey, V., Huber, W. & Hahne, F. Genefilter: methods for filtering genes from high-throughput experiments. R package version 1 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol 15 https://doi.org/10.1186/s13059-014-0550-8 (2014).
van der Maaten, L. & Hinton, G. Visualizing Data using t-SNE. J Mach Learn Res 9, 2579–2605 (2008).
Google Scholar
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550, https://doi.org/10.1073/pnas.0506580102 (2005).
Article ADS CAS PubMed PubMed Central Google Scholar
Park, M., Cha, S. & Yi, J.-M. Digital transformation of herbal medicine: Conversion to biological entity data using tonifying herbal medicine-induced transcriptome sequencing_Tonifying_A549, https://identifiers.org/GEO:GSE244687 (2023).
Park, M., Cha, S. & Yi, J.-M. Digital transformation of herbal medicine: Conversion to biological entity data using tonifying herbal medicine-induced transcriptome sequencing_Tonifying_HepG2, https://identifiers.org/GEO:GSE244707 (2023).
Park, M., Cha, S. & Yi, J.-M. Digital transformation of herbal medicine: Conversion to biological entity data using tonifying herbal medicine-induced transcriptome sequencing_Tonifying_HT29, https://identifiers.org/GEO:GSE244694 (2023).
Park, M., Cha, S. & Yi, J.-M. Digital transformation of herbal medicine: Conversion to biological entity data using tonifying herbal medicine-induced transcriptome sequencing_Tonifying_SW1783, https://identifiers.org/GEO:GSE245912 (2023).
Park, M. et al. KORE-map_Sample quality, https://doi.org/10.6084/m9.figshare.26019445.v1 (2024).
Park, M. et al. KORE-map_Sequencing quality, https://doi.org/10.6084/m9.figshare.26020525.v1 (2024).
Download references
Total RNA isolation and RNA sequencing were conducted by LAS Co. Ltd., South Korea. This study was supported by Grant number KSN1722122 from the Korea Institute of Oriental Medicine.
These authors contributed equally: Musun Park, Sang-Min Park, Haeseung Lee.
Korean Medicine (KM) Data Division, Korea Institute of Oriental Medicine, Daejeon, Republic of Korea
Musun Park & Seongwon Cha
College of Pharmacy, Chungnam National University, Daejeon, Republic of Korea
Sang-Min Park
College of Pharmacy, Pusan National University, Busan, Republic of Korea
Haeseung Lee
KM Application Center, Korea Institute of Oriental Medicine, Daegu, Republic of Korea
Aeyung Kim
KM Convergence Research Division, Korea Institute of Oriental Medicine, Daejeon, Republic of Korea
No Soo Kim, Yu Ri Kim & Jin-Mu Yi
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
You can also search for this author in PubMed Google Scholar
All authors contributed to the study. M.P. and J.Y. were responsible for data generation. M.P., S.P., and H.L. analyzed the data. M.P. uploaded and curated data. S.P. and H.L. visualized the data and performed formal analysis. M.P., S.P., H.L., J.Y. and S.C. drafted the manuscript. All authors reviewed and edited the manuscript.
Correspondence to Jin-Mu Yi or Seongwon Cha.
The authors declare no competing interests.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
Reprints and permissions
Park, M., Park, SM., Lee, H. et al. KORE-Map 1.0: Korean medicine Omics Resource Extension Map on transcriptome data of tonifying herbal medicine. Sci Data 11, 974 (2024). https://doi.org/10.1038/s41597-024-03734-x
Download citation
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03734-x
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
Advertisement
© 2025 Springer Nature Limited
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.







